Scientists (Georgios Mikaelian, Haralambos Sarimveis, Doros Theodoru) from the School of Chemical Engineering from National Technical University of Athens (NTUA), were lead- and co-authoring in the following publication:
Mikaelian, G.; Sarimveis, H.; Theodorou, D. N.; & Megariotis, G. (2025). Thermodynamics and Kinetics of the Deintercalation of a Novel Anthracycline from Double-Stranded Oligonucleotide DNA. The Journal of Physical Chemistry B, 129, 8335–8350. https://doi.org/10.1021/acs.jpcb.5c03021
Mikaelian et al. perform an in silico investigation of the thermodynamics and kinetics of berubicin deintercalation from double-stranded DNA oligonucleotides using well-tempered metadynamics (WT-MetaD), an enhanced sampling technique that allows detailed exploration of molecular free-energy landscapes. Berubicin – a novel anthracycline currently in phase II clinical trials for glioblastoma multiforme – is distinct among anthracyclines in its ability to cross the blood–brain barrier, making it a particularly promising chemotherapeutic compound. The study extends the authors’ previous work (J. Phys. Chem. B 2024, 128, 6291–6307), which focused on the binding thermodynamics of the berubicin–DNA intercalated complex; here, they address the unbinding (deintercalation) process in mechanistic and kinetic detail.
The authors stress that “a mere thermodynamic study of the binding and unbinding process is not sufficient,” since drug efficacy depends not only on equilibrium affinity but also on kinetic stability. As they note, “deintercalation is considered a process of tremendous significance and its kinetic analysis is directly related to the pharmacological efficacy of drugs.” Their computational framework quantifies both aspects through free-energy surface (FES) mapping and transition-state analysis within the WT-MetaD formalism.
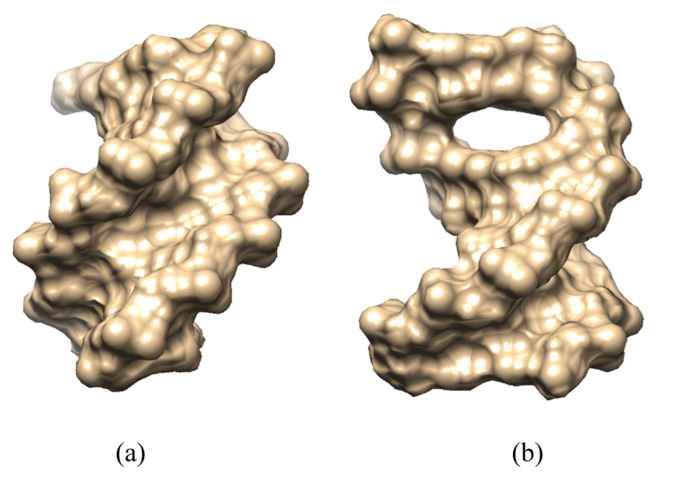
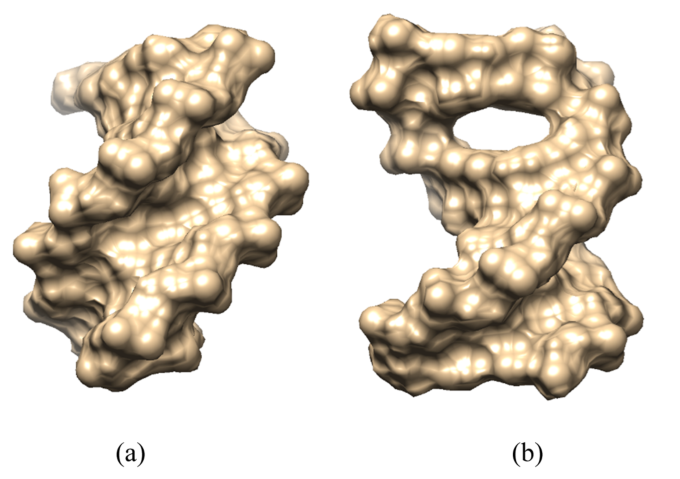
Visualization of 5′-d(ACGTAC|GT)-3′ when free in water (a) and when berubicin is bound to its intercalation site (b). The oligonucleotides are depicted as molecular surfaces using the CHIMERA software (figure taken from the publication)
Two DNA sequences – 5′-d(ACGTAC|GT)-3′ and 5′-d(TGT|ACA)-3′ – were simulated in explicit water, each bound to protonated berubicin at physiological pH (7.2). The molecular dynamics simulations, carried out with GROMACS 2021.4 interfaced with PLUMED 2.7.2, employed ~100–150 ns trajectories. The collective variables (CVs) describing deintercalation were a distance (X) between the drug and the intercalation site and an angle (θ) representing the relative orientation of berubicin’s anthraquinone plane to the DNA helix.
Through the WT-MetaD bias potential, the system was driven out of deep free-energy wells corresponding to bound states, yielding Gibbs free-energy surfaces as functions of X and θ. This analysis identified three relevant conformational basins: (1) the intercalated state, (2) a reshuffling intermediate, and (3) the minor-groove-bound state. According to the authors, “our simulations reveal that the deintercalation mechanism involves two stable states, the intercalated and the minor groove-bound state, as well as a reshuffling state. The rate-limiting step is the transition from the intercalated to the reshuffling state.”
The calculated Gibbs free-energy surfaces displayed two pronounced minima, corresponding to the intercalated and minor groove-bound states, separated by a high barrier plateau associated with the reshuffling transition. Quantitative results showed standard binding Gibbs energies (ΔG°) of approximately −60 kJ mol⁻¹ for the intercalated and −35 kJ mol⁻¹ for the groove-bound configurations, in strong agreement with their earlier double-decoupling results and with experimental data on related anthracyclines such as doxorubicin and daunorubicin.
The minor-groove-bound state was found to be thermodynamically less favorable but kinetically significant, as it forms part of the multi-step release pathway. This hierarchy aligns with experimental observations that “binding Gibbs energies for the minor groove-bound state fall in the range of −24 to −42 kJ mol⁻¹, lower than those of the intercalated state.”
Using transition-state theory applied to WT-MetaD trajectories, the authors determined deintercalation rate constants and mean residence times. The rate-limiting step, the transition from intercalated to reshuffling, exhibited activation Gibbs energies of 70–76 kJ mol⁻¹, indicative of substantial structural rearrangement. Corresponding mean residence times (τ) were 1.07 s for 5′-d(ACGTAC|GT)-3′–berubicin and 0.10 s for 5′-d(TGT|ACA)-3′–berubicin at 310 K, increasing markedly at lower temperatures. These values are comparable to or longer than those of clinically used anthracyclines, suggesting high kinetic stability and, potentially, sustained pharmacological activity.
The longer τ observed for the 5′-d(ACGTAC|GT)-3′ sequence was attributed to stronger electrostatic and stacking interactions, a higher base-pairing enthalpy, and more effective hydration shielding of the intercalation site. The authors demonstrate that “water molecules play an important role during the deintercalation process,” where invasion of solvent into the binding pocket triggers unbinding, an effect consistent with mechanistic models of protein, ligand dissociation.
A comprehensive structural analysis revealed that helical parameters (twist and rise) and base-pair overlap values change markedly during deintercalation. In the intercalated state, the DNA helix is locally distorted (rise ≈ 7 Å; twist ≈ 160°), while in the groove-bound and unbound states, it relaxes to canonical B-DNA geometry. The intermediate reshuffling state exhibited partial collapse of the intercalation cavity, confirming a drug-induced cavity formation mechanism rather than spontaneous opening of intercalation sites. Energetically, van der Waals forces dominate in the intercalated state, while electrostatic and hydrogen-bond interactions gain prominence in the groove-bound state. The computed hydrophobic contributions to the binding free energy (ΔG_hpb ≈ −40 to −50 kJ mol⁻¹) underline the key role of nonpolar stabilization, in line with previous anthracycline studies.
The study elucidates, at atomistic resolution, the multi-step mechanism of berubicin unbinding from DNA, offering quantitative thermodynamic and kinetic parameters that align with and extend experimental understanding. The authors conclude that “the first stage of deintercalation is rate-limiting and fully determines the kinetic stability of the complex.” Their findings indicate that berubicin exhibits deintercalation times equal to or exceeding those of established anthracyclines, which may contribute to its potent and durable pharmacological action against glioblastoma.
Methodologically, the work showcases WT-MetaD as a powerful tool for dissecting slow biomolecular transitions, establishing a benchmark for computational pharmacokinetics of DNA-binding drugs. The mechanistic model combining intercalated → reshuffling → minor-groove-bound → unbound states provides a unified picture of anthracycline–DNA dynamics and supports rational design of future CNS-permeable anticancer agents.
Follow this link to read the full publication.
Parts of the research of this work (NTUA) has been funded by the European Union`s R&I project PINK (grant agreement # 101137809).