Scientists (Nikoletta‑Maria Koutroumpa, Maria Antoniou, Iseult Lynch, Christoforos Kyprianou, Andreas Tsoumanis, Haralambos Sarimveis, and Antreas Afantitis) from the PINK project partners NovaMechanics Ltd. (NOVA), School of Chemical Engineering, National Technical University of Athens (NTUA), and School of Geography, Earth and Environmental Sciences, University of Birmingham (UoB), were lead- and co-authoring in the following publication:
Koutroumpa, N.-M., Antoniou, M., Varsou, D.-D., Papavasileiou, K. D., Sidiropoulos, N. K., Kyprianou, C., Tsoumanis, A., Sarimveis, H., Lynch, I., Melagraki, G., & Afantitis, A. (2025). Titania: an integrated tool for in silico molecular property prediction and NAM-based modeling. Molecular Diversity, 29, 3555–3573. https://doi.org/10.1007/s11030-025-11196-5
This publication introduces Titania, a comprehensive and freely accessible web-based platform for in silico molecular property prediction, embedded within the Enalos Cloud Platform. As the authors state, “Titania features an intuitive, user-friendly interface, allowing researchers, regardless of computational expertise, to easily employ models for property prediction of novel compounds”. The work integrates computational toxicology, cheminformatics, and artificial intelligence into a single environment that adheres to OECD and European Commission standards for model validation and transparency.
Titania represents an advanced implementation of Quantitative Structure–Property Relationship (QSPR) and Quantitative Structure–Toxicity Relationship (QSTR) modeling, which seeks to correlate molecular structure with physicochemical and biological properties. The authors emphasize that “efficient and reliable prediction of molecular properties is crucial for rational compound design in the chemical industry”, and that these models enable researchers to “predict the properties of untested chemicals prior to synthesis, using only their molecular structure information”.
The authors developed predictive models for nine fundamental physicochemical and biological endpoints, namely: octanol/water partition coefficient (logP), water solubility (logS), hydration free energy in water (Free-Solv), vapor pressure (logVP), boiling point (BP), cytotoxicity, mutagenicity, blood–brain barrier (BBB) permeability, and bioconcentration factor (logBCF). The datasets used to train and validate the models were compiled from established databases such as MoleculeNet, EPA EPI Suite, and PubChem BioAssay, all curated to eliminate duplicates and structural ambiguities.
The modeling process employed machine learning (ML) algorithms including k-nearest neighbors (kNN), random forest (RF), support vector machines (SVM), and multi-layer perceptron (MLP). Following OECD guidelines, each model was extensively validated through leave-one-out (LOO), fivefold cross-validation, external blind testing, and Y-randomization. The reported performance was robust: “Q² values ranged from 0.69 to 0.89 for regression models, and accuracies ranged from 0.787 to 0.864 for classification models”.
All developed models were “documented and validated according to OECD guidelines, ensuring transparency, reliability, and regulatory compliance”, and further supported by QMRF (QSAR Model Reporting Format) documentation. Importantly, the models define a domain of applicability (DoA) based on Euclidean distance metrics to “filter out unreliable results (extrapolated predictions)”, an essential component for ensuring reproducibility and regulatory acceptance. A major achievement of this work lies in the seamless integration of the models into the Titania web tool ( https://enaloscloud.novamechanics.com/EnalosWebApps/titania/). Built on the ZK Framework and Java/JavaScript technologies, Titania allows researchers to upload or draw molecular structures, compute multiple property predictions simultaneously, and visualize molecules in 3D using an “NGL viewer with multiple atom representation styles and annotation features”.
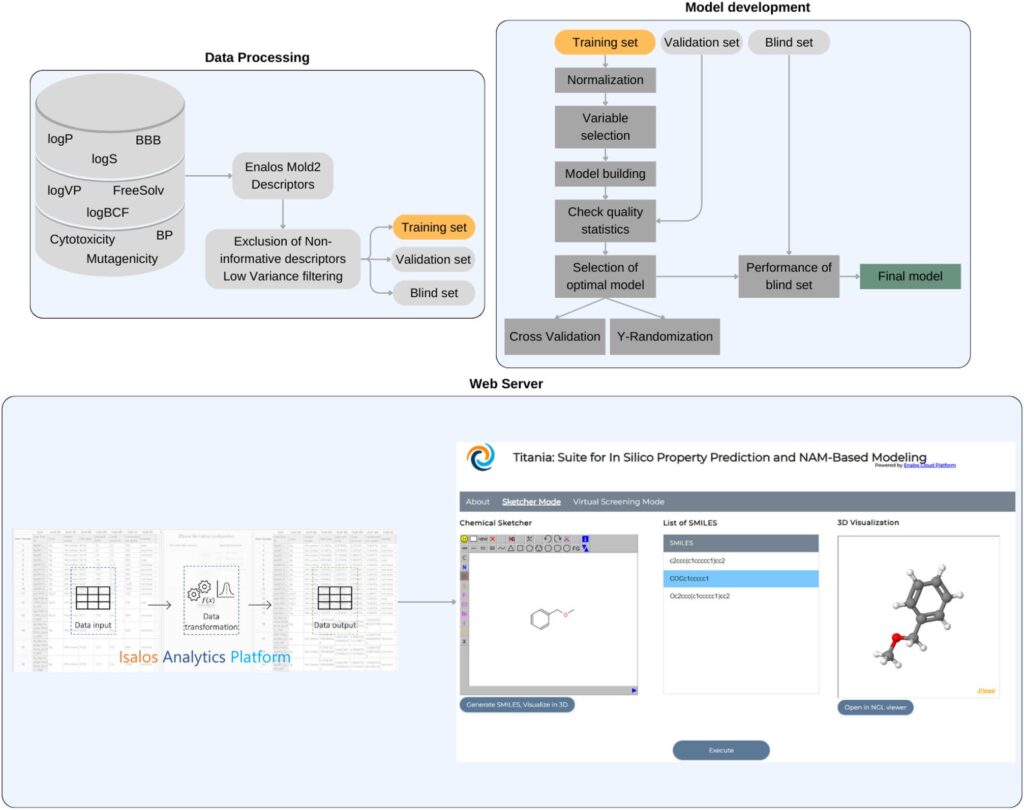
Schematic representation of the development and use of the Titania models. Molecular featurization, data pre-processing, and model development were performed on KNIME, and models were transferred into the Isalos Analytics Platform. The developed models were easily deployed through the Enalos Cloud Platform as a userfriendly web-service called Titania.
Titania’s innovation extends to transparency and interpretability—features often missing in comparable tools. Each prediction is accompanied by color-coded reliability indicators based on the model’s DoA and provides “the k-nearest neighbors and their distances from the input substances, allowing evaluation of structural similarity patterns”. This explicit visualization of the predictive rationale represents a key methodological advancement toward explainable AI in cheminformatics.
The system is further integrated with the ChemPharos database, ensuring open access to the curated datasets used in model training. As the authors note, “all of the datasets, including the compounds’ descriptors and properties/toxicity, used to develop the QSPR/QSTR models are made available through the ChemPharos database”. This commitment to data FAIRness—Findable, Accessible, Interoperable, and Reusable—is a defining feature of the Titania framework.
The paper positions Titania within the ecosystem of established ADMET prediction tools, including SwissADME, ADMETlab 3.0, pkCSM, and ADMET-AI, concluding that “Titania excels in QSAR model transparency, providing QMRF documentation for all models, model interpretability… and applicability domain assessment”. While the other tools emphasize speed or endpoint breadth, Titania prioritizes regulatory compliance and interpretability, making it particularly relevant for New Approach Methodologies (NAMs) and non-animal testing strategies.
Three case studies demonstrate the practical utility of Titania: (i) prediction of physicochemical and BBB permeability parameters for approved drugs; (ii) comparative property analysis for per- and polyfluoroalkyl substances (PFAS) to support safer chemical substitution; and (iii) toxicity and permeability assessment of Endoplasmic Reticulum Aminopeptidase 1 (ERAP1) inhibitors as potential therapeutic candidates. In each case, Titania successfully identified “similar compounds exhibiting properties closely aligned with the query compound” and enabled mechanistic interpretation via nearest-neighbor analysis.
Titania is positioned as a next-generation in silico modeling environment that unites machine learning, data FAIRness, and regulatory transparency under the same platform. The authors assert that “Titania, and the underpinning Enalos Cloud Platform, aims to become a valuable tool in drug discovery and material research, supporting the reduction of in vitro experiments”. By embedding validated QSPR/QSTR models into a cloud-based graphical interface, Titania operationalizes OECD-aligned predictive modeling, advancing the field toward fully digital, mechanistically interpretable NAMs. Its originality lies in bridging scientific rigor and accessibility: a platform where users can perform high-throughput, interpretable virtual screening while maintaining full regulatory traceability and adherence to FAIR data principles.
Follow this link to read the full publication.
Parts of the research of this work (NovaM, NTUAS, and UoB) has been funded by the European Union`s R&I project PINK (grant agreement # 101137809).